-
Notifications
You must be signed in to change notification settings - Fork 14
Motivation Behind EpiRust
"The greatest shortcoming of the human race is our inability to understand the exponential function." - Dr. Albert Allen Bartlett
Each country poses different challenges in face of a pandemic, and India has its own - Scale of geography, population, and economy; coupled with diversity and connectedness. Policy makers need to rely, not only on their own expertise and experience but also to be more collaborative and creative in charting a response.
Epidemic response for COVID19 pandemic on a global scale is an interdisciplinary challenge. Disciplines such as epidemiology, immunology, medicine, pharmaceuticals and equipment manufacturing, economics, statistics, engineering, and public health policy are all tied together in search for an appropriate and rapid response.
An overarching aspect across these functional areas is optimization: Is a response plan feasible that minimizes damage and fits into given time, budget, and people? Usually, such an exploration needs a set of options to choose from for each concern, and stitching the choices together to construct a plan.
Information science and technology can help the scientists and the policy makers by developing mathematical models of an epidemic and their computational simulations of various what-if and if-what scenarios. Going by the famous quote of George P. Box, “All models are wrong, some are useful.”, the quest is to develop models useful for policy makers.
Even simple mathematical models offer important insights into disease spread.
These models are called as Compartmentalized Models, in that an individual moves from one compartment to another; from susceptible to infected to recovered in this SIR model (figure 3). Other similar models are Susceptible-Exposed-Infected-Recovered (SEIR), Susceptible-Infected-Susceptible (SIS), and so on.

However, these differential equations based models have many short-comings, and a critical one is that they fail to model how an epidemic actually spreads. A disease spreads via contacts, either by an infected person, substance, or by an intermediary (a vector in epidemiology jargon) such as mosquitoes. Such interactions have boundaries in that people must come together, or touch infected substances, which cannot be modeled by differential equations.
To address these problems, scientists are developing relatively new methods such as ‘agent-based’ or ‘individual-based’ models (here an agent is a virtual person in a virtual world). These methods are becoming increasingly popular, for example, the United States’ Center for Disease Control used insights generated by such models to help control the 2014 West Africa Ebola Outbreak. In India, we are yet to see their adoption at this scale, partly because these methods are computationally far more intensive, and need an interdisciplinary collaboration between computer science and epidemiology to build a body of knowledge and implementations that is more suited for the Indian context. For computer scientists, these methods offer many high quality computational challenges in terms of modeling and execution efficiency.
ThoughtWorks’ Engineering for Research initiative (E4R) being interested in such challenges started exploring agent-based modelling for large-scale epidemiological simulations sometime around the mid-2019, with a goal to develop ideas and implementations around efficient, flexible and robust simulation models.

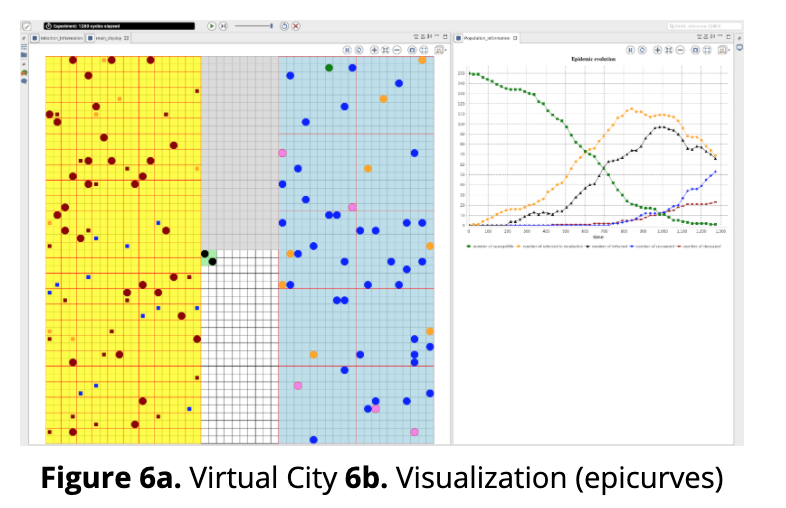
In the second half of 2019, we at E4R developed an agent-based simulation to model spread of smallpox disease, which could spread over a virtual population of up to 10,000 agents. We modeled each agent living its own life (figure 6a) such as following the daily routine of home activities including sleep (yellow area), commutation (grey area), office work (blue area), and so on. For commutation, each agent has an option to take either public or private transportation, and when such choices are made, it alters the probability of catching a disease. Once caught, the infected person can pass to others, which may not happen immediately due to incubation period as per the model. To contain the spread of a disease, many interventions are modeled; for example, mass vaccination, quarantine or isolation, and so on. We used a framework namely GAMA for this development. To help understand the dynamics of a simulation run, we added visualization (see figure 6b) which are supported by GAMA.
However, we felt the overall simulation was slower than our anticipation. For 10,000 agents, the simulation took approximately 60 minutes to complete on a 128-core server using all the 128 cores. To achieve our target to simulate a large population representing a city like Pune, we decided to implement our own framework ground-up using the Rust programming language.
We have named it the ‘EpiRust’ framework.
After re-implementation of the core disease model and its spread in a population, simulation run time for 10,000 agents came down from 60 minutes to a little over 20 seconds, that too using just one CPU core.
In addition, we now have completed a city-lockdown intervention and are now modeling a healthcare system with hospitals and staff. We have modeled a representative population of Pune city based on census data. And yes, we have modeled a guesstimated disease dynamics for COVID-19, with a set of parameters which an epidemiologist can alter.
We have released it in open source, so that some of the experts we have been talking to, can also make use of it in their own pursuit. We are already in discussion with experts in epidemiology and related disciplines in order to take this work to policy makers.
The source code is available on the GitHub repository of EpiRust.